I am looking for a post-doc for the NAABB project in the lab. The announcement is below, please spread the word to all interested parties.....
The USDA-ARS Plant Genetics Research Unit at the Danforth Plant Sciences Center in St Louis, MO is seeking a Postdoctoral Research Associate, for a two year appointment with possibility for extension. Ph.D. in plant sciences, genetics, molecular biology or related field is required. Salary is commensurate with experience ($57,408-$74,628) plus benefits. Citizenship restrictions apply. The incumbent will conduct research studying the systems biology of algal mineral nutrition. A successful candidate will combine bioinformatics, microbiology and genetics to understand how interactions between elements and gene-element interactions affect the growth of biofuel-relevant strains of Algae. Send application materials and references to Dr. Ivan Baxter at ivan.baxter@ars.usda.gov . The position is open until filled. USDA/ARS is an equal opportunity employer and provider.
Monday, August 23, 2010
Thursday, June 17, 2010
Arabidopsis RILs paper is published
Our paper on "Natural Genetic Variation in Selected Populations of Arabidopsis thaliana Is Associated with Ionomic Differences" has just been published in PLoS ONE. I'm really happy that this paper is out because it demonstrates something that we have been talking about for several years now: that the ionome is dynamic and interconnected, and the relationships between elements are dependent on both genetics and the environment. The first three figures in this paper describe experiments which were the basis for three other papers that we have already published. Here is the abstract and none technical summary:
Abstract:
Controlling elemental composition is critical for plant growth and development as well as the nutrition of humans who utilize plants for food. Uncovering the genetic architecture underlying mineral ion homeostasis in plants is a critical first step towards understanding the biochemical networks that regulate a plant's elemental composition (ionome). Natural accessions of Arabidopsis thaliana provide a rich source of genetic diversity that leads to phenotypic differences. We analyzed the concentrations of 17 different elements in 12 A. thaliana accessions and three recombinant inbred line (RIL) populations grown in several different environments using high-throughput inductively coupled plasma- mass spectroscopy (ICP-MS). Significant differences were detected between the accessions for most elements and we identified over a hundred QTLs for elemental accumulation in the RIL populations. Altering the environment the plants were grown in had a strong effect on the correlations between different elements and the QTLs controlling elemental accumulation. All ionomic data presented is publicly available at www.ionomicshub.org.
Non-technical Summary:
Understanding how plants regulate element composition of tissues is critical for agriculture, the environment, and human health. Sustainably meeting the increasing food and biofuel demands of the planet will require growing crops with fewer inputs such as the primary macronutrients phosphorus (P) and potassium (K). Ionomics is the study of elemental accumulation in living systems using high-throughput elemental profiling. With this technique, we can rapidly generate large quantities of data on thousands of samples, allowing for the profiling of large genetic mapping populations and the discovery of hundreds of loci important for elemental accumulation. We have used this approach to sample the natural diversity present in collections of a model plant, the wild mustard Arabidopsis, and mapping populations derived from those collections. We find that when the soil environment changes, the identity of the genes important for elemental accumulation changes as well. We also find that elements will have different relationships between them depending on the environment and the lines under study. This suggests that crop varieties developed for improved elemental uptake and accumulation will be highly environment specific.
Abstract:
Controlling elemental composition is critical for plant growth and development as well as the nutrition of humans who utilize plants for food. Uncovering the genetic architecture underlying mineral ion homeostasis in plants is a critical first step towards understanding the biochemical networks that regulate a plant's elemental composition (ionome). Natural accessions of Arabidopsis thaliana provide a rich source of genetic diversity that leads to phenotypic differences. We analyzed the concentrations of 17 different elements in 12 A. thaliana accessions and three recombinant inbred line (RIL) populations grown in several different environments using high-throughput inductively coupled plasma- mass spectroscopy (ICP-MS). Significant differences were detected between the accessions for most elements and we identified over a hundred QTLs for elemental accumulation in the RIL populations. Altering the environment the plants were grown in had a strong effect on the correlations between different elements and the QTLs controlling elemental accumulation. All ionomic data presented is publicly available at www.ionomicshub.org.
Non-technical Summary:
Understanding how plants regulate element composition of tissues is critical for agriculture, the environment, and human health. Sustainably meeting the increasing food and biofuel demands of the planet will require growing crops with fewer inputs such as the primary macronutrients phosphorus (P) and potassium (K). Ionomics is the study of elemental accumulation in living systems using high-throughput elemental profiling. With this technique, we can rapidly generate large quantities of data on thousands of samples, allowing for the profiling of large genetic mapping populations and the discovery of hundreds of loci important for elemental accumulation. We have used this approach to sample the natural diversity present in collections of a model plant, the wild mustard Arabidopsis, and mapping populations derived from those collections. We find that when the soil environment changes, the identity of the genes important for elemental accumulation changes as well. We also find that elements will have different relationships between them depending on the environment and the lines under study. This suggests that crop varieties developed for improved elemental uptake and accumulation will be highly environment specific.
Sunday, April 25, 2010
Digestion!
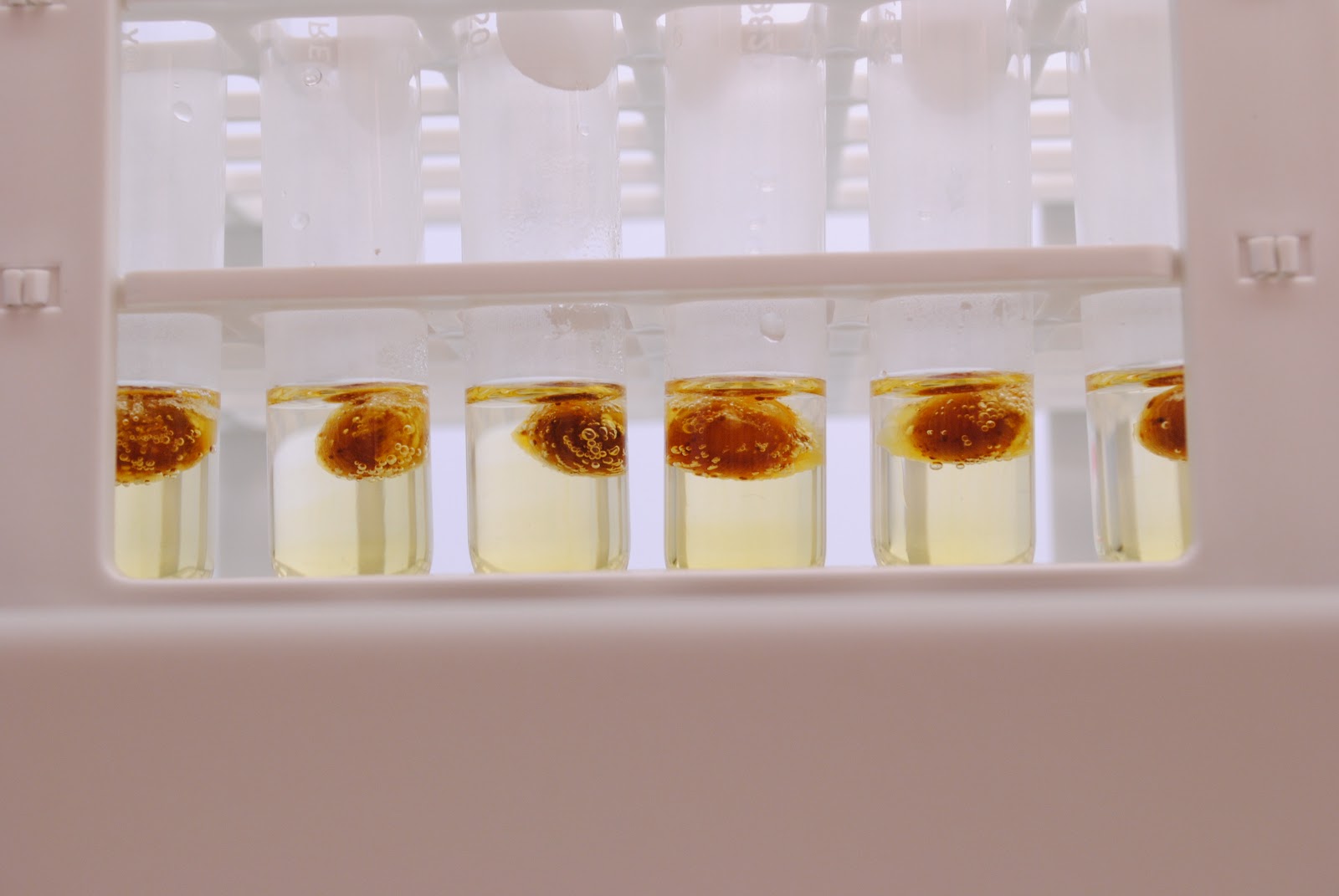
What you are looking at are individual soybeans digesting in Nitric acid. This process breaks down almost every molecule in the bean to its elemental components, which allows us to quantify the levels of each element within the sample using ICP-MS. The renovations to the lab are finally complete and we are starting to work out the kinks in the process. Pretty soon we will be able to start working our way through the backlog of samples that have accumulated in our lab.
Arabidopsis Association Paper published in Nature
The big Nature paper, to which we contributed phenotype data, is now out. Its a great example of the power of plant genetics, many different phenotypes evaluated on a common population resulting in a wealth of really interesting associations. Its way cool, but the bigger populations that we have analyzed are even better and soon we will be submitting some really cool new results for publication.
Saturday, April 17, 2010
Introducing Walter
How cool is Ionomics? So cool that we are able to recruit beer scientists to the cause! Introducing Walter Iverson, the newest member of the Baxter lab.

Walter has years of analytical chemistry expirience and will be running the ICPs and the elemental profiling facility. Welcome Walter!

Walter has years of analytical chemistry expirience and will be running the ICPs and the elemental profiling facility. Welcome Walter!
Friday, February 19, 2010
Getting really, really, close.....
We are getting really close to having a functional ionomics lab. Yesterday, the last major piece of equipment, the fan for the top of the exhaust system was installed on the roof. As you can see, it was quite a production....

Almost all the internal construction is finished and we are on schedule for the ICPs to be installed at the beginning of March.
In other news, the weighing robot is almost finished. This week I visited Paul Armstrong at the USDAs Engineering and Wind Erosion Research Unit in Manhattan Kansas. Paul has built a robot that can take single corn and soybean seeds from a 48 well plate, weigh them and then put them in a specified digestion tube. This turns out to be one of the bottleneck steps in our sample prep, so the robot will save us hours every day. Here is a photo of the robot with Paul in the background....

and here is a front view......

Before you know it, we will be cranking through samples.


Almost all the internal construction is finished and we are on schedule for the ICPs to be installed at the beginning of March.
In other news, the weighing robot is almost finished. This week I visited Paul Armstrong at the USDAs Engineering and Wind Erosion Research Unit in Manhattan Kansas. Paul has built a robot that can take single corn and soybean seeds from a 48 well plate, weigh them and then put them in a specified digestion tube. This turns out to be one of the bottleneck steps in our sample prep, so the robot will save us hours every day. Here is a photo of the robot with Paul in the background....

and here is a front view......

Before you know it, we will be cranking through samples.
Monday, February 1, 2010
Maize Grant Submitted
In collaboration with Owen Hoekenga (Boyce Thompson Institute/USDA-ARS/Cornell University), Mourad Ouzzani (Purdue University), Margaret Smith (Cornell University), and Paul Anderson (Danforth Center) I just submitted a grant to the NSF-Plant Genome Research Program entitled "Mineral Nutrient Gene Discovery and Gene X Environment Interactions Using the Nested Association Mapping Population in Maize".
Here is the abstract:
Maize is the most widely adapted and adopted crop on the planet. This is largely due to the amazing degree of genetic and phenotypic diversity that can be harnessed into adaptation to local conditions. While progress has been made in some aspects of adaptation, e.g. flowering time, little progress has been made with respect to adaptation to soil conditions at the molecular and genetic levels. This is ironic given the importance of plant-soil interactions as they relate to agricultural efficiency, sustainability and productivity. We will utilize the Nested Association Mapping (NAM) population, a unique and powerful genetic resource, to identify genes controlling the elemental composition (the ionome) of maize grain. We will measure the levels of 20 different elements: P, Ca, S, K, Mg, Sr, Rb (macronutrients or their chemical analogs); B, Cu, Fe, Zn, Mn, Co, Ni, Mo (micronutrients of significance to plant and human health); Na, Al, As, Se and Cd (minerals causing agricultural or environmental problems). We will leverage grain samples from the 5,000 recombinant inbred lines that constitute the NAM population, which have already been grown at four different locations with widely different soils. One expected outcome of our project is the identification, at single gene resolution, of loci and alleles that alter the accumulation of the mineral nutrients and toxic elements from different soil conditions. We will confirm these results and identify potential causative polymorphisms by association analysis. For 20 selected loci, we will create Heterogeneous Inbred Families (HIFs) that an extended team of collaborators will help us to evaluate in multiple soil environments, which we will select based upon screening soil samples provided by our extended team. The HIFs will confirm the predicted allelic affects and allow us investigate the interactions between genetic and environmental factors to determine grain quality. Additional outcomes for our project will be the identification of hundreds of genetic loci and dozens of nucleotide polymorphisms that determine the mineral nutritional content of maize grain. We will also gain a better understanding of how many of these genes interact with environmental factors.
Here is the abstract:
Maize is the most widely adapted and adopted crop on the planet. This is largely due to the amazing degree of genetic and phenotypic diversity that can be harnessed into adaptation to local conditions. While progress has been made in some aspects of adaptation, e.g. flowering time, little progress has been made with respect to adaptation to soil conditions at the molecular and genetic levels. This is ironic given the importance of plant-soil interactions as they relate to agricultural efficiency, sustainability and productivity. We will utilize the Nested Association Mapping (NAM) population, a unique and powerful genetic resource, to identify genes controlling the elemental composition (the ionome) of maize grain. We will measure the levels of 20 different elements: P, Ca, S, K, Mg, Sr, Rb (macronutrients or their chemical analogs); B, Cu, Fe, Zn, Mn, Co, Ni, Mo (micronutrients of significance to plant and human health); Na, Al, As, Se and Cd (minerals causing agricultural or environmental problems). We will leverage grain samples from the 5,000 recombinant inbred lines that constitute the NAM population, which have already been grown at four different locations with widely different soils. One expected outcome of our project is the identification, at single gene resolution, of loci and alleles that alter the accumulation of the mineral nutrients and toxic elements from different soil conditions. We will confirm these results and identify potential causative polymorphisms by association analysis. For 20 selected loci, we will create Heterogeneous Inbred Families (HIFs) that an extended team of collaborators will help us to evaluate in multiple soil environments, which we will select based upon screening soil samples provided by our extended team. The HIFs will confirm the predicted allelic affects and allow us investigate the interactions between genetic and environmental factors to determine grain quality. Additional outcomes for our project will be the identification of hundreds of genetic loci and dozens of nucleotide polymorphisms that determine the mineral nutritional content of maize grain. We will also gain a better understanding of how many of these genes interact with environmental factors.
Subscribe to:
Posts (Atom)